May 28, 21 · The Cas9 nuclease widely used for genome editing is derived from natural bacterial defense systems that protect against invading viruses Cas9 is directed by RNA guides to cut matching viral DNAMar 24, 21 · Several previous studies show that mismatches in the seed region (the 10–12 base pairs adjacent to the PAM) are critical and determine the specificity of Cas9 compared to the distal part of the sgRNA sequence (nonseed sequence) (HsuCas9 will only cleave a given locus if the gRNA spacer sequence shares sufficient homology with the target DNA Once the Cas9gRNA complex binds a putative DNA target, the seed sequence (810 bases at the 3′ end of the gRNA targeting sequence) will begin to anneal to the target DNA If the seed and target DNA sequences match, the gRNA will continue to anneal to the target DNA in a

Quantification Of Cas9 Binding And Cleavage Across Diverse Guide Sequences Maps Landscapes Of Target Engagement Science Advances
Cas9 seed region
Cas9 seed region-Nov 02, 15 · The CRISPR/Cas9 system has been developed in recent years for genome editing, and it has been rapidly and widely adopted by the scientific community 1 The RNAguided enzyme Cas9 originates from the CRISPRCas adaptive bacterial immune systemJan 01, 15 · The sequence of the seed region determines the frequency of a "seed NGG" in the genome, and controls the effective concentration of the Cas9sgRNA complex (Cas9 binding or sgRNA abundance and specificity) 21, 25 Meanwhile, Urich seeds are likely to result in decreased sgRNA abundance and increased specificity since multiple U's in the sequence can induce



Figure 3 From Crispr Interference Crispri For Sequence Specific Control Of Gene Expression Semantic Scholar
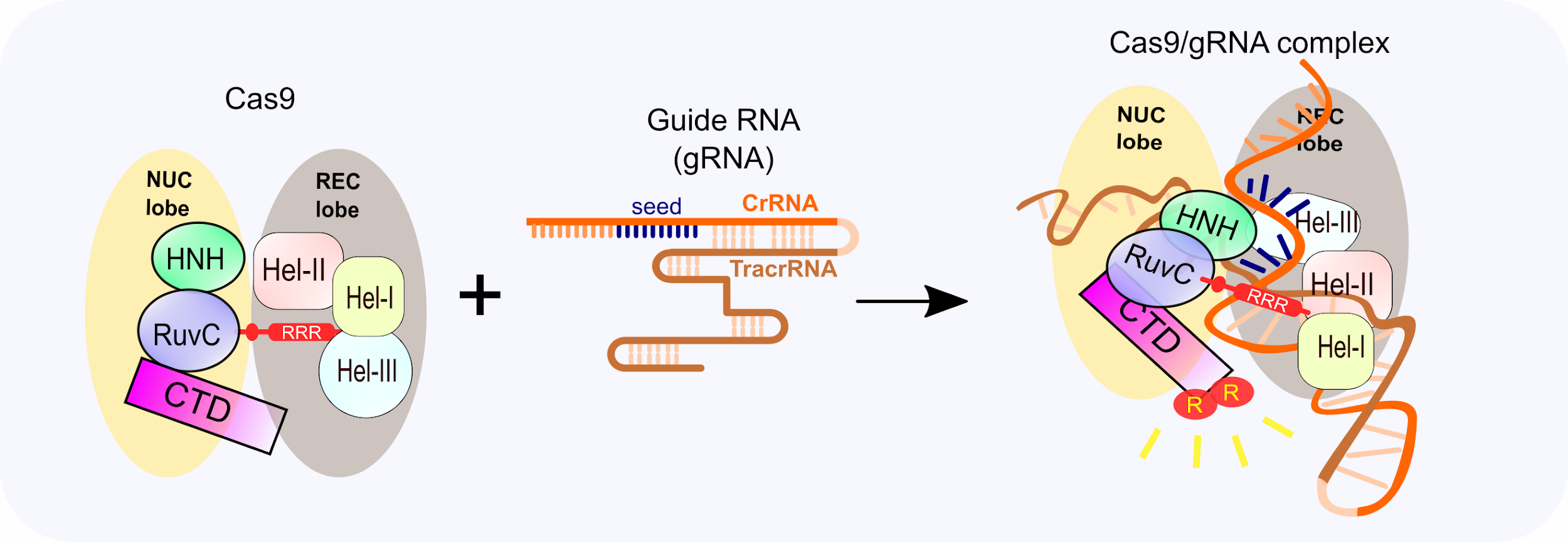
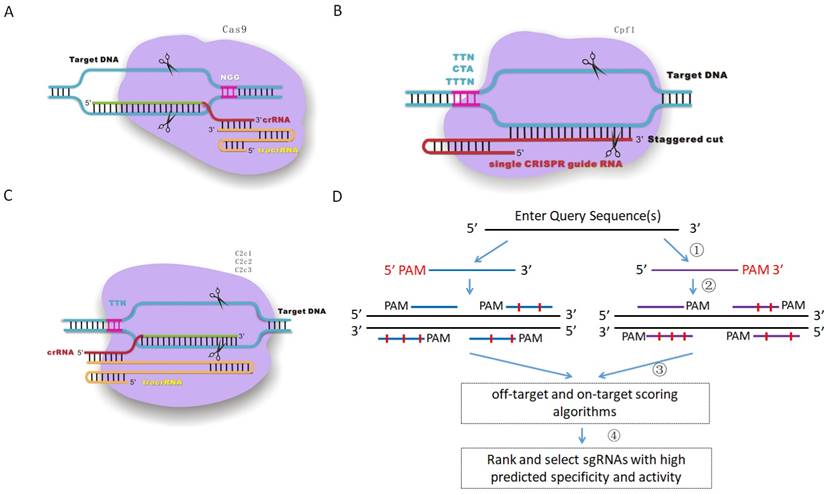
Jun 12, 19 · This is consistent with the observation that the mismatch of the twocrRNA nucleotide adjacent to PAM is intolerable while offtarget DSB could occur in the DNA sequence homology to the seed region Figure 2 PAMbinding of Cas9/gRNA complex and initiation of crRNA/target DNA hybridizationJul 16, · These essential 13 nt of the spacer region have been dubbed the 'seed sequence'8,23 Genomic regions with incomplete homology to the spacer region which contain all or most of the seed sequence could be targeted by the Cas9, resulting in offtarget effects24 Detection and prevention of this offtarget activity is essentialThe nucleotides in the seed region are recognized by Cas9 protein via the arginine residues in the bridge helix and the REC1 domains within the REC lobe This seed region serves as a sensitive element for precise recognition and cleavage of the nucleotide DNA sequence within the target genomic locus 11
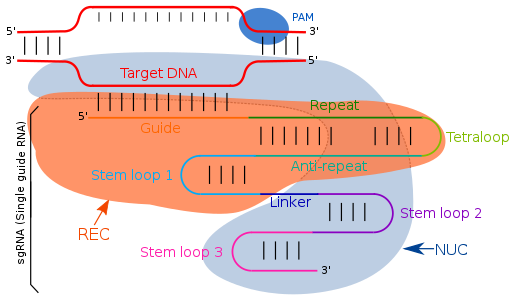
Jun 26, 15 · We propose that the preordered PAM recognition region of the Cas9sgRNA complex initiates DNA interrogation, followed by base pairing between a short PAMproximal segment of DNA (1 or 2 bp) and theAt the molecular level, Cas9 associates with the guide RNA (either the naturally occurring crRNAtracrRNA duplex structure or an engineered sgRNA) in a binary protein–RNA complex that uses a nucleotide region at the 5′ end of the guide RNA to locate a matching sequence in a dsDNA target (Jinek et al, 12;Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications Its main function is to cut DNA and thereby alter a cell's genome The CRISPRCas9 genome editing technique was a significant
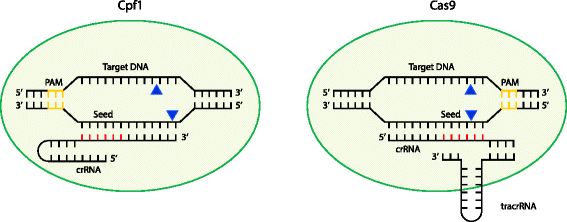
Mismatches within the PAMdistal region (positions 11) are better tolerated than mismatches within the seed region, the Cas9RNP can still bind sites with some seed region mismatches More recently, highcontent kinetic studies of Cas9RNP binding in vitro have shown that mismatches in the PAMproximal region dissociate more readily than distal mismatches, but that distalFig 1 Focusing on the interaction between the seed region of gRNA and Cas9 protein (a) Interactive sites between the OH and the residues of Cas9 revealed by the crystal structure The numbering starts from the 50end of gRNA with a standard nt guide sequenceNov 17, 15 · Note that the guide RNA in Cas9 is an RNA duplex involving crRNA and transactivating CRISPR RNA (tracrRNA), whereas Cpf1 uses a single crRNA Upon sufficient complementarity in the seed region (red), Cpf1 and Cas9 nucleases will make two singlestranded cuts (blue triangles) resulting in a doublestranded break



Crispr Plant


A Benchmark Of Computational Crispr Cas9 Guide Design Methods
CRISPR/Cas9 has become one of the most promising techniques for genome editing in plants and works very well in poplars with an Agrobacteriummediated transformation systemWe selected twelve genes, including SOC1, FUL, and their paralogous genes, four NFPlike genes and TOZ19 for three different research topics The gRNAs were designed for editing, and, together with aCas9sgRNABinary and SeedPaired Nme1Cas9sgRNAdsDNA Ternary Complexes (A) Domain organization of Nme1Cas9 (B) Schematic representation of the 135nt sgRNA are located in the seed region, whereas nucleotides 1–14 are disordered This observation suggests that in the absence of target DNA, the Nme1Cas9 sgRNA seed is preordered in aMar 08, 18 · The crystal structure of Cas9–sgRNA indicates that RNA at the seed region (ten nucleotides at the 3′ end of the guide sequence) is essential for Cas9–sgRNA binding and recognition of targeted DNA



Factors That Impact Cas9 Speci Fi City Top Before Cas9 Is Introduced Download Scientific Diagram


Sgrnacas9 A Software Package For Designing Crispr Sgrna And Evaluating Potential Off Target Cleavage Sites
Cas9 normally tolerates mismatches that are located at the 5′ end of the gRNA, outside the seed region This is supported in our studies, as we found that a single nucleotide mutation located outside the crRNA seed region did not affect the portability of our in vitro assembled Cas9Sternberg et al 14) Cas9 and its derivatives offer a repertoire of functions Wildtype Cas9 nuclease sequence, also referred to as the seed region The seed region has been defined as the sequence of 6 to 12bp immediately upstream of to the PAM siteAug 03, 18 · This result implies substantial reversibility during Rloop formation—a late transition state—and defies common descriptions of a 'seed' region"


Frontiers A New Age In Functional Genomics Using Crispr Cas9 In Arrayed Library Screening Genetics



Crispr Cas9 Abm Inc
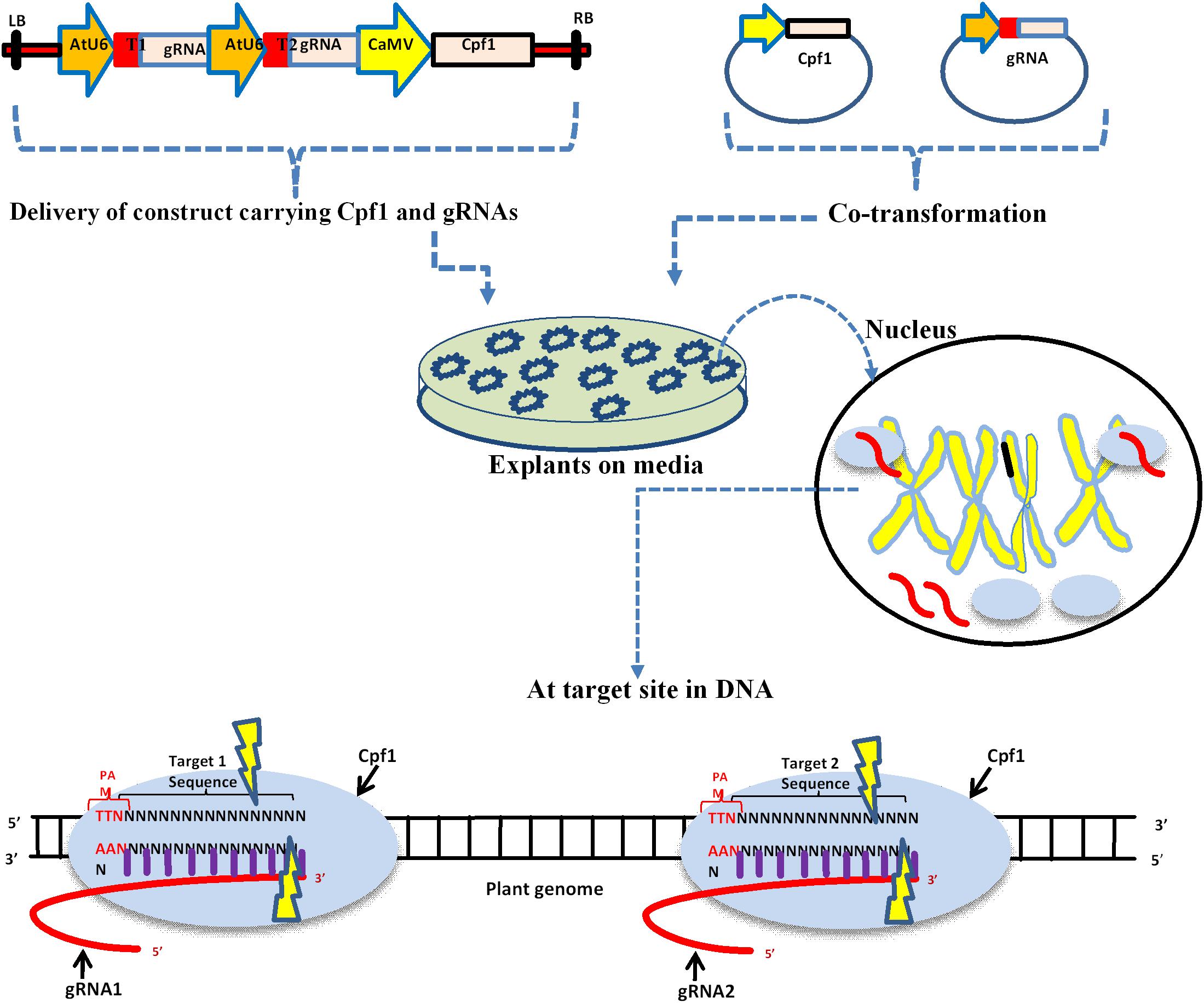
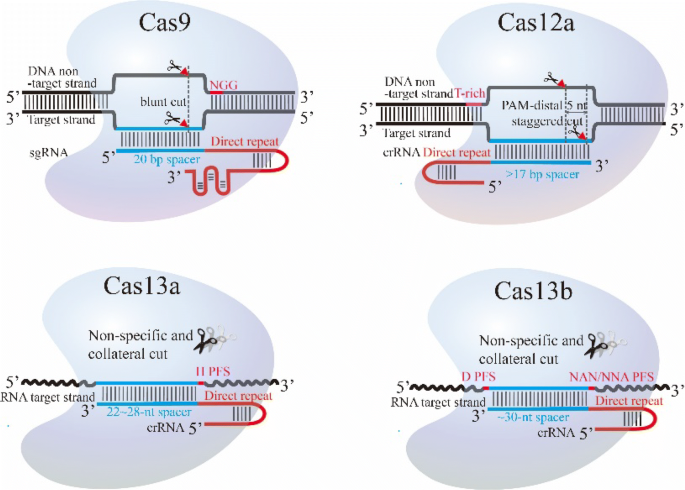
Cas9 technology to introduce sequencespecific deleterious point mutations at the eIF(iso)4E locus in Arabidopsis thaliana to suc bases in the seed region (12 consecutive nucleotides upstream of the PAM), as these parameters have been experimentally validated as the minimum requirements for directing Cas9 to cleaveFeb 04, · The CRISPR/Cas9 system is a powerful tool for targeted gene editing in many organisms including plants However, most of the reported uses of CRISPR/Cas9 in plants have focused on modifying one or a few genes, and thus the overall specificity, types of mutations, and heritability of gene alterations remain unclear Here, we describe the molecular characterizationMar 10, 17 · Experiments suggest that the nt long protospacer can be divided into two regions, the seed (PAMproximal) region within 10 base pairs from the PAM and the nonseed (PAMdistal) region with 10



Patterns Of Crispr Cas9 Activity In Plants Animals And Microbes Bortesi 16 Plant Biotechnology Journal Wiley Online Library


Crispr Systems Doudna Lab
Jun 03, · Red indicates the seed region that is less tolerance to mismatches for Cas9 binding b The flowchart of the bioinformatics analysis procedure to evaluate gRNA specificities The guide sequences of gRNA that target rice mitochondrial and chloroplast 16S rRNA gene (mtgRNA and cpgRNA) were extracted according to the requirements for Cas9/gRNAContacts between Cas9 and the seed region of the crRNA deform the seed region from BIBC 100 at University of California, San DiegoCRISPR/Cas9 System Step 1 Provide a genomic region to search for candidate CRISPR targets Input a DNA sequence in FASTA format ?



Off Target Genome Editing Wikipedia



Cas9 Cuts And Consequences Detecting Predicting And Mitigating Crispr Cas9 On And Off Target Damage Newman Bioessays Wiley Online Library
Jun 03, 21 · It is known that mismatches within the socalled "seed region" of the protospacer comprising 10 nucleotides proximal to the PAM site, are more likely to abolish target cleavage Therefore, it is tempting to hypothesize that the single mismatch in the cyp1a3 region targeted by our crRNA was not tolerated by the CRISPR/Cas9 system and didThe bacterial CRISPR/Cas9 system (Clustered Regularly Interspaced Palindromic Repeats associated protein 9 system) is among the youngest players in the gene editing playground Ensure the 12nt 'seed' region is adjacent to the PAM (Protospacer adjacent motif)Jan 01, 19 · Tilapia miRNA125 was selected as the target to examine whether mutation could be induced in the seed region using CRISPR/Cas9 gRNA containing restriction enzyme Mse I was designed in the seed sequence of miRNA125 Coinjection of gRNA and Cas9 mRNA led to indels formation in the seed region


Frontiers The Rise Of The Crispr Cpf1 System For Efficient Genome Editing In Plants Plant Science


Guide Rna Wikipedia
Feb 19, 21 · Such work has revealed that the Cas9 ribonucleoprotein (RNP) first associates to an NGG protospacer adjacent motif (PAM) and then hybridizes to 8 to 12 target nucleotides abutting the PAM known as the "seed" regionPerfect complementarity between the seed region of sgRNA and target DNA is necessary for Cas9mediated DNA targeting and cleavage, whereas imperfect base pairing at the nonseed region is much more tolerated for target binding specificity RNA–DNA HeteroduplexApr 18, 18 · The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPRassociated nuclease 9 (Cas9) genome editing technology provides such a tool, enabling revolutionary advances in both arenas of plant biology CRISPR/Cas systems are bacterial adaptable immune systems against foreign DNA sources such as bacteriophages or plasmids ( Wiedenheft



Addgene Crispr Guide



Identification Of Guide Intrinsic Determinants Of Cas9 Specificity The Crispr Journal
Unlike previous engineering of gRNA that generally focused on the RNA part only but neglected RNA–protein interactions, we aimed at the interactive sites between 2′OH of ribose in the seed region of gRNA and Cas9 protein and identified that modifications of 2′OH at specific sites could be utilized to regulate the Cas9 activityCas9 Nuclease, S pyogenes, is an RNAguided endonuclease that catalyzes sitespecific cleavage of double stranded DNA The location of the break is within the target sequence 3 bases from the NGG PAM (Protospacer Adjacent Motif) The PAM sequence, NGG, must follow the targeted region on the opposite strand of the DNA with respect to the region complementary sgRNA sequenceHere, we optimized CRISPR interference (CRISPRi) for use in Caulobacter Using Streptococcus thermophilus CRISPR3 or Streptococcus pasteurianus CRISPR systems, we show that the coexpression of a catalytically dead form of Cas9 (dCas9) with a single guide RNA (sgRNA) containing a seed region that targets the promoter region of a gene of interest


The Cpf1 Crispr Cas Protein Expands Genome Editing Tools Genome Biology Full Text


The Magic Cut On Target Dna By Crispr Cas9 Bioscope
Oct 30, 19 · In addition, despite the high specificity of the CRISPRCas9 system, offtarget mutations can occur at sites that have sequence similarity to the target site 24,25,26, especially when there are no mismatches in the PAMproximal 8–12 nucleotide 'seed region' of the guide sequence 27,28,29,30 Such offtarget sites may be present in nontarget genes or nontargetMay 22, 14 · NGG and is a requirement for Cas9 binding to its target region (Jinek et al 12;Karvelis et al, 13) Watson–Crick base pairing between the guide sequence



Crispr Cas9 Guide Rna Specificity


Crispr Offinder A Crispr Guide Rna Design And Off Target Searching Tool For User Defined Protospacer Adjacent Motif
Cas9 is a DNA endonuclease with two active domains (red triangles) cleaving each of the two DNA strands three nucleotides upstream of the PAM The five nucleotides upstream of the PAM are defined as the seed region for target recognition Applications of CRISPRCas9Mar 02, 21 · Here, we established the seed fluorescence reporter (SFR)assisted CRISPR/Cas9 systems in maize (Zea mays L), using the red fluorescent DsRED protein expressed in the endosperm (EnSFR/Cas9), embryos (EmSFR/Cas9), or both tissues (Em/EnSFR/Cas9) All three SFRs showed distinct fluorescent patterns in the seed endosperm and embryo thatNov 25, 16 · Following PAM recognition, Cas9 helicase activity promotes target DNA unwinding and the progressive invasion of the nucleotide guide segment of the Cas9bound sgRNA, beginning with the sgRNA "seed" region at the 3′ end of the guide segment



Synthetic Crispr Rna Cas9 Guided Genome Editing In Human Cells Pnas


Class 2 Crispr Cas An Expanding Biotechnology Toolbox For And Beyond Genome Editing Cell Bioscience Full Text
Oct 02, 14 · The tolerance of Cas9 binding and cleavage activities for a small number of mismatches in the 5′distal portion of the target relative to its seed region was further explored by assaying a series of probes containing alternating 2 or 3 base pair (bp) mismatches across this area While binding efficiencies were somewhat consistent for each ofMay 01, 17 · Genomewide characterization of Cas9 binding by chromatin immunoprecipitation and highthroughput sequencing has shown binding but little cleavage at targets with seed region mutations (17, 18) as well as substantially decreased Cas9 binding in some cases (18, 19) Seed mutations are thus expected to severely compromise drive copy, and for ourFeb 01, 21 · Our maximal productive binding measurement instead appears to align with the conventional understanding of Cas9 targets, which have an 8 to 10bp seed region that is sensitive to disruption, an 8 to 11bp PAMdistal region that is largely resilient, and an intermediate zone sensitive to large perturbations


Crispr Offinder A Crispr Guide Rna Design And Off Target Searching Tool For User Defined Protospacer Adjacent Motif



Figure 3 From Crispr Interference Crispri For Sequence Specific Control Of Gene Expression Semantic Scholar
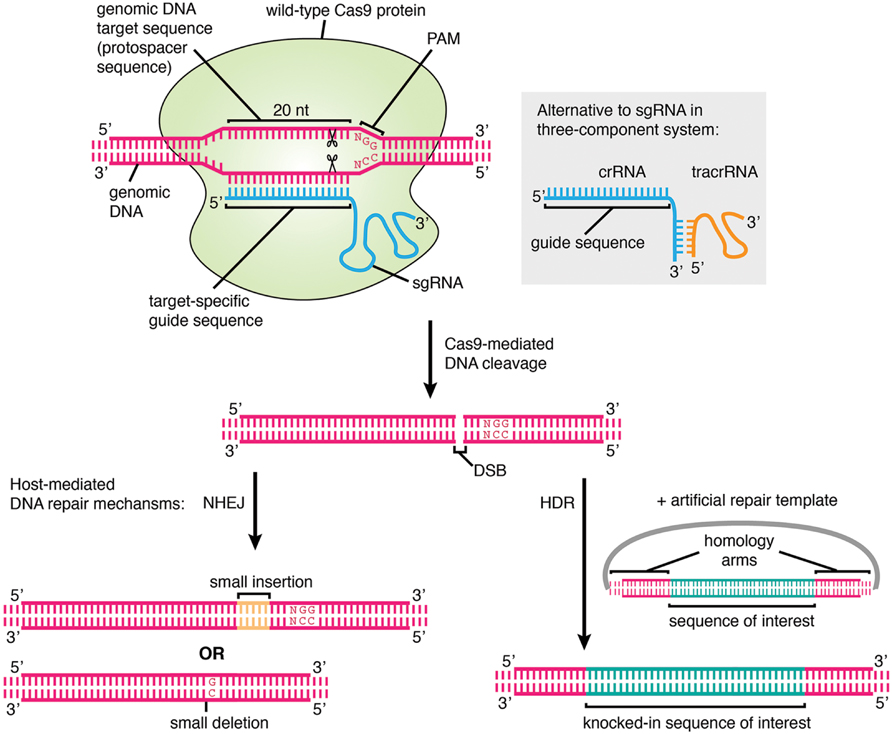
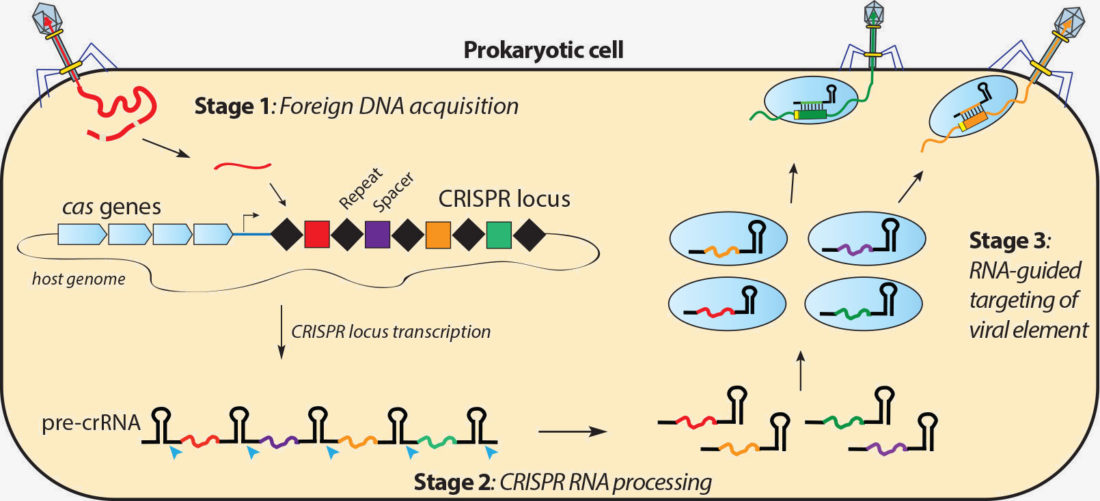
CRISPR (/ ˈ k r ɪ s p ər /) (which is an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote They are used to detect and destroy DNA from similarThe action mechanism of CRISPR/Cas9 System As shown in figure 1, the PAM sequence causes recognition of the Cas9 protein, allowing the singlestranded guide RNA linked to the tracrRNA to recognize the target sequence site, ensuring that the Cas9 protein and the genome stably binding, causing cleavage of the target site (usually the 3 and 4



High Frequency Off Target Mutagenesis Induced By Crispr Cas9 In Arabidopsis And Its Prevention By Improving Specificity Of The Tools Biorxiv



Crispr Cas9 Abm Inc


Crispr Cpf1 Proteins Structure Function And Implications For Genome Editing Cell Bioscience Full Text


Frontiers Crispr Cas9 Genome Editing Technology A Valuable Tool For Understanding Plant Cell Wall Biosynthesis And Function Plant Science



Addgene Crispr Guide



Design Principle Of A Crispr Cas9 Expression Vector For Construction Of Download Scientific Diagram



Nucleosomes Selectively Inhibit Cas9 Off Target Activity At A Site Located At The Nucleosome Edge Journal Of Biological Chemistry



Crispr Cas Systems In Genome Editing Methodologies And Tools For Sgrna Design Off Target Evaluation And Strategies To Mitigate Off Target Effects Manghwar Advanced Science Wiley Online Library


Off Target Genome Editing Wikipedia



The Crispr Cas9 System For Plant Genome Editing And Beyond Sciencedirect



One Or Two Mismatches In The Seed Can Be Tolerated For Crispr Activity Download Scientific Diagram


The Biology Of Native And Adapted Crispr Cas Systems Journal Of Young Investigators



Off Target Effects In Crispr Cas9 Mediated Genome Engineering Sciencedirect



A Programmable Dual Rna Guided Dna Endonuclease In Adaptive Bacterial Immunity Science
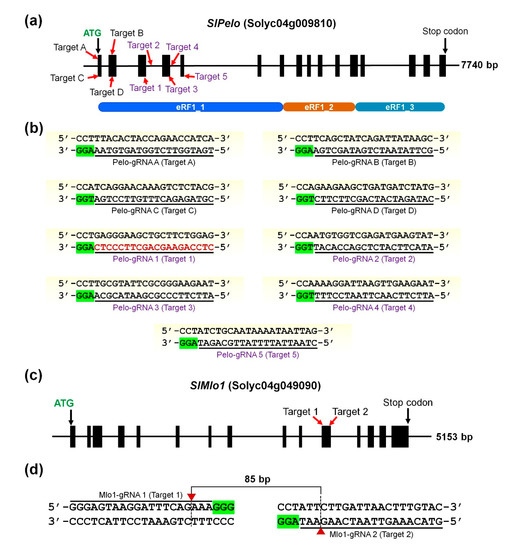


Ijms Free Full Text Crispr Cas9 Mediated Generation Of Pathogen Resistant Tomato Against Tomato Yellow Leaf Curl Virus And Powdery Mildew Html


A Cas9 Is Directed To A Specific Genomic Target By The First Nt Of Download Scientific Diagram



Protein Engineering Strategies To Expand Crispr Cas9 Applications



A Model For Cas9 Target Binding And Cleavage A In The Unbound State Download Scientific Diagram



High Efficiency Targeting Of Non Coding Sequences Using Crispr Cas9 System In Tilapia G3 Genes Genomes Genetics



Crispr Diagnosis And Therapeutics With Single Base Pair Precision Trends In Molecular Medicine
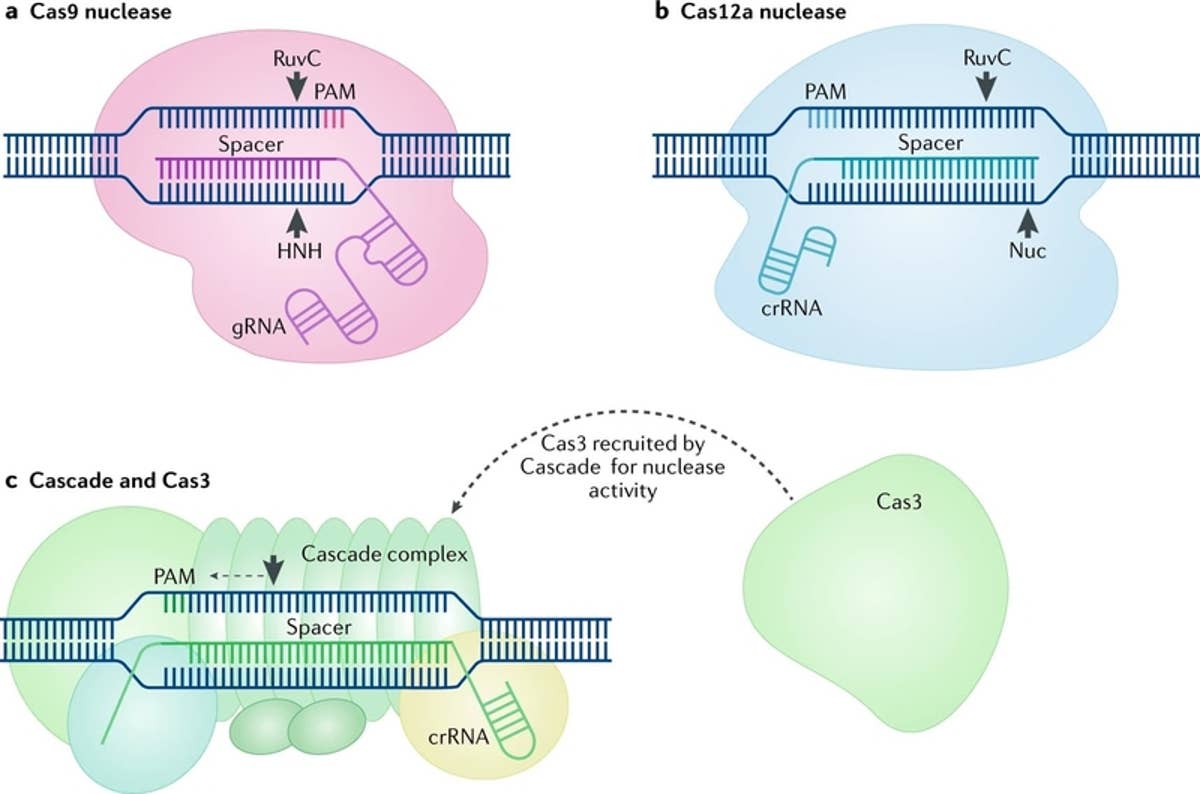


How To Choose The Right Cas9 Variant For Every Crispr Experiment
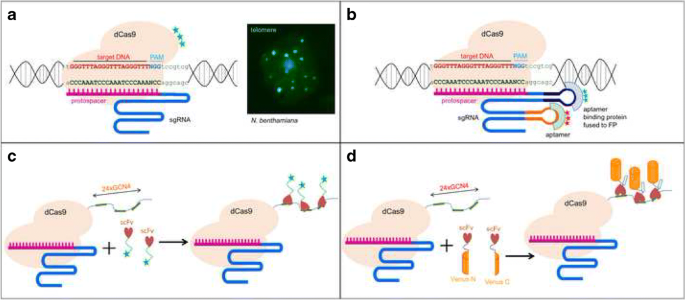


Application And Prospects Of Crispr Cas9 Based Methods To Trace Defined Genomic Sequences In Living And Fixed Plant Cells Springerlink


Team Cu Boulder Project Modeling 14 Igem Org



Interference By Clustered Regularly Interspaced Short Palindromic Repeat Crispr Rna Is Governed By A Seed Sequence Pnas
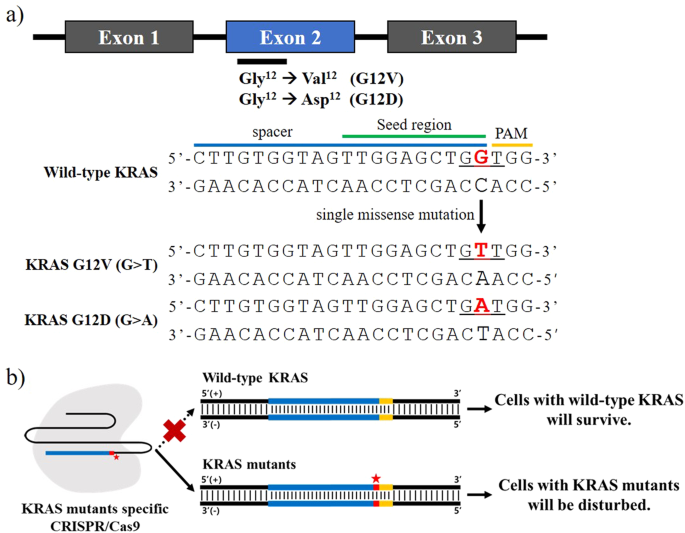


Selective Targeting Of Kras Oncogenic Alleles By Crispr Cas9 Inhibits Proliferation Of Cancer Cells Scientific Reports
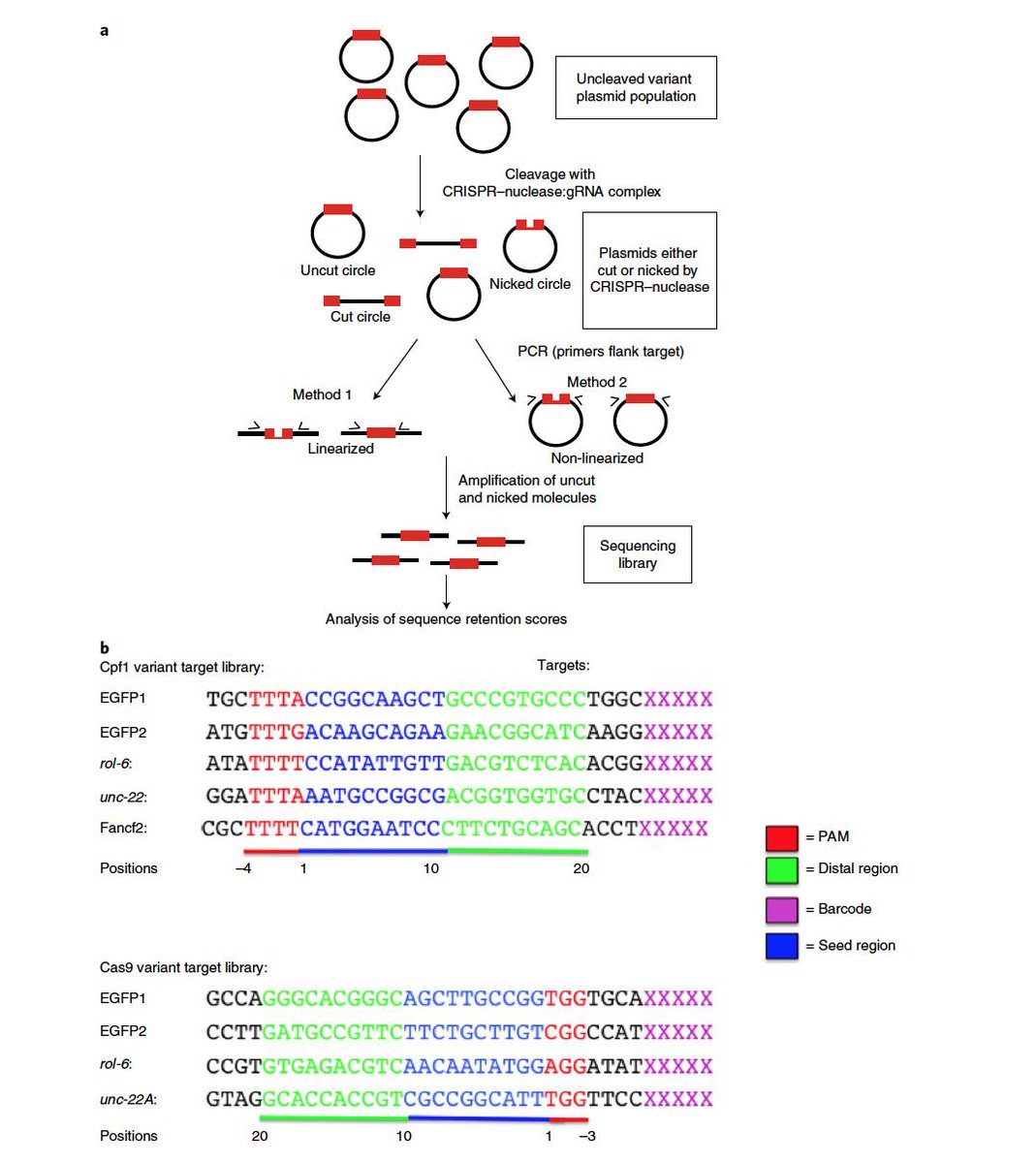


Dr Gaetan Burgio Md Phd Auf Twitter Interesting This Paper Published Today In Naturemicrobiol Shows That Crispr Cas9 Or Cas12a Cpf1 Can Display An Inherent Potent Nicking Activity In Yeast This Nicking
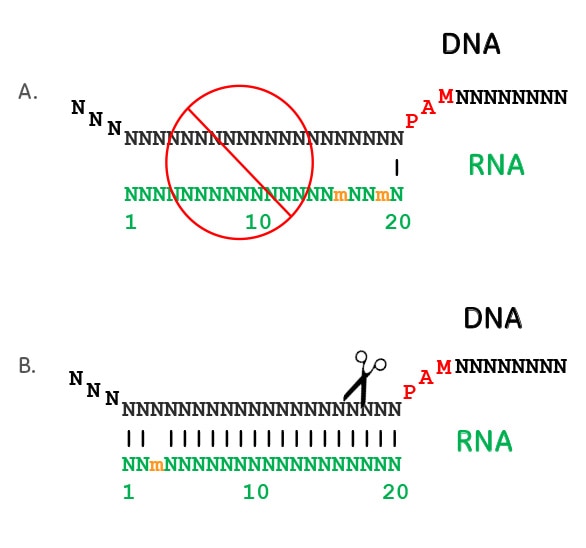


Crispr Cas9 Guide Rna Specificity
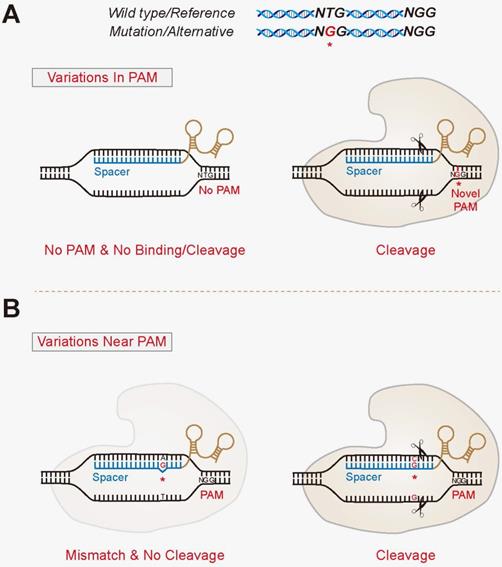


Allele Specific Genome Targeting In The Development Of Precision Medicine
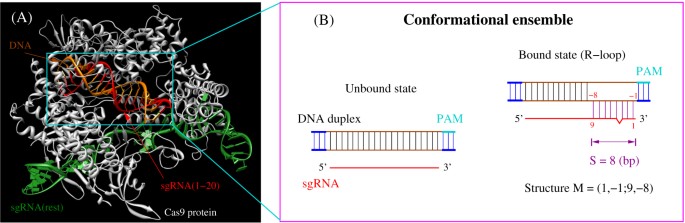


Crispr Cas9 Cleavage Efficiency Correlates Strongly With Target Sgrna Folding Stability From Physical Mechanism To Off Target Assessment Scientific Reports



High Efficiency Targeting Of Non Coding Sequences Using Crispr Cas9 System In Tilapia G3 Genes Genomes Genetics



Quantification Of Cas9 Binding And Cleavage Across Diverse Guide Sequences Maps Landscapes Of Target Engagement Science Advances



Crispr Cas9 Mediated Genome Editing Increases Lifespan And Improves Motor Deficits In A Huntington S Disease Mouse Model Molecular Therapy Nucleic Acids



Schematic Illustration Of The Crispr Cas9 System Structure And The Download Scientific Diagram



Engineered Rna Interacting Crispr Guide Rnas For Genetic Sensing And Diagnostics The Crispr Journal


Full Article Defining The Seed Sequence Of The Cas12b Crispr Cas Effector Complex
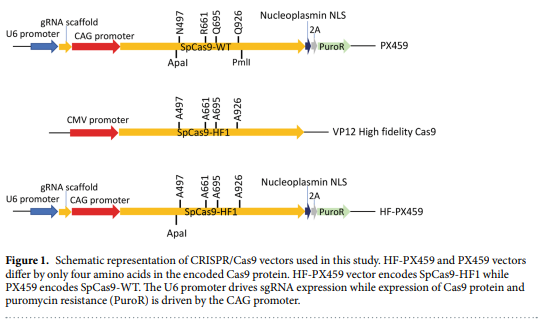


High Fidelity Crispr Cas9 Increases Precise Monoallelic And Biallelic Editing Events In Primordial Germ Cells Engormix
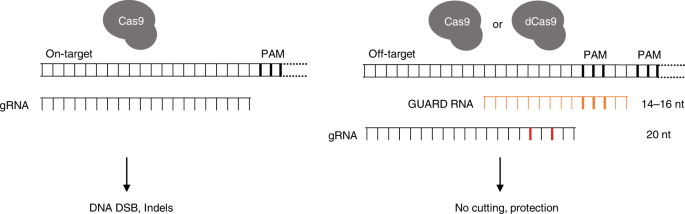


Crispr Guard Protects Off Target Sites From Cas9 Nuclease Activity Using Short Guide Rnas Nature Communications
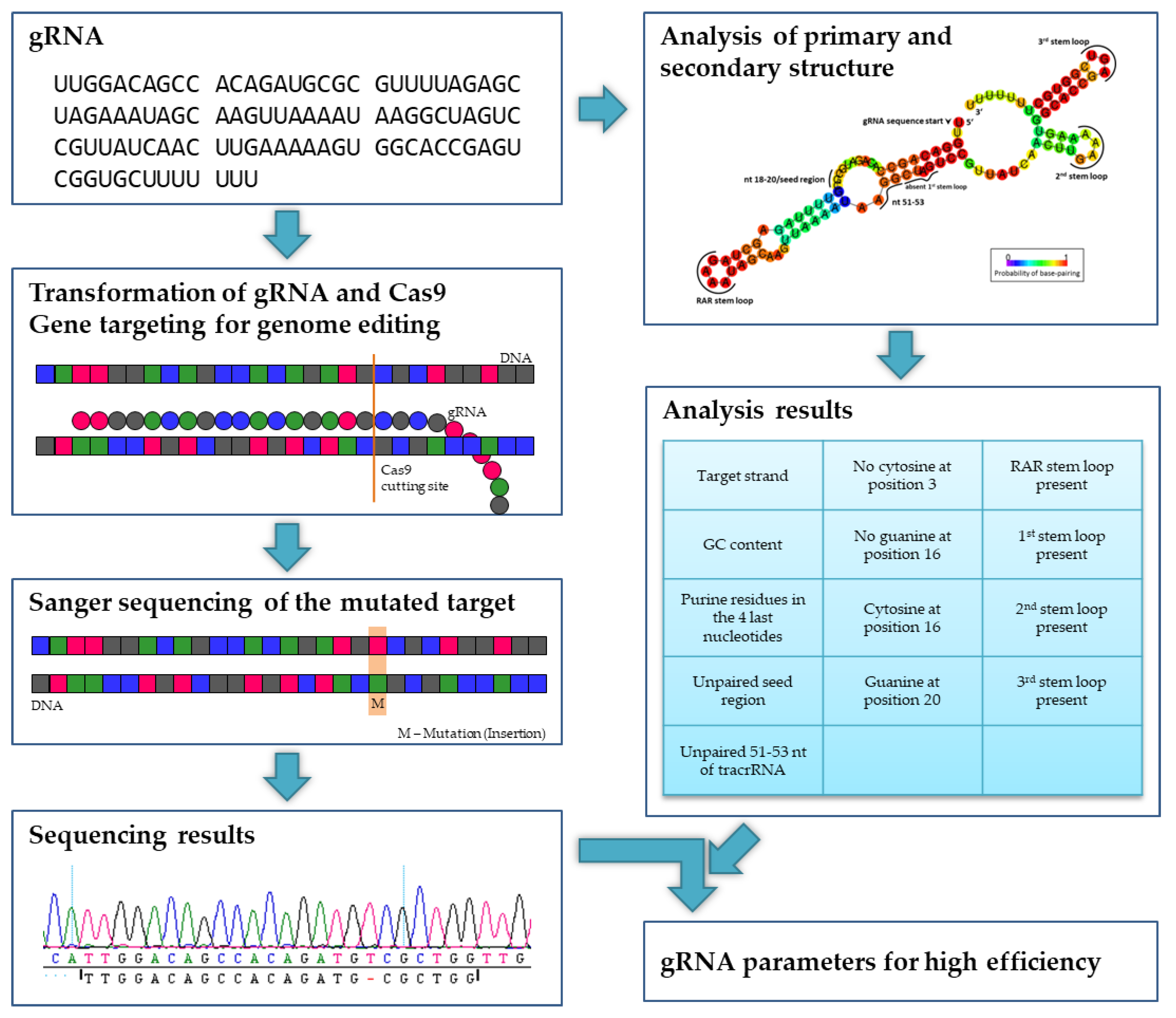


Ijms Free Full Text Evaluating The Efficiency Of Grnas In Crispr Cas9 Mediated Genome Editing In Poplars Html



Target Specificity Of The Crispr Cas9 System Abstract Europe Pmc
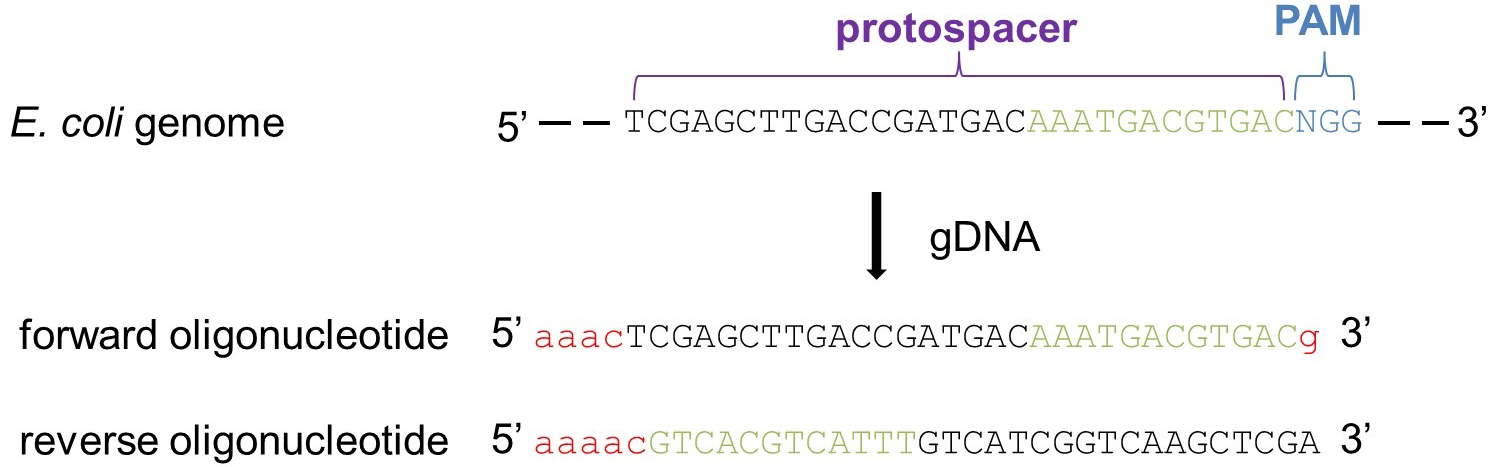


使用crispr Cas9在大肠埃希杆菌中进行逐步多基因敲除 Bio Protocol
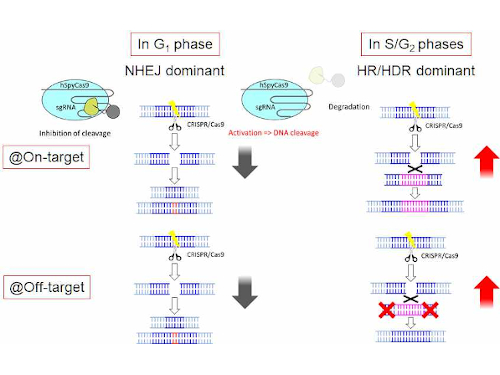


Seedquest Central Information Website For The Global Seed Industry



Enhancement Of Target Specificity Of Crispr Cas12a By Using A Chimeric Dna Rna Guide Biorxiv


Crispr Cas
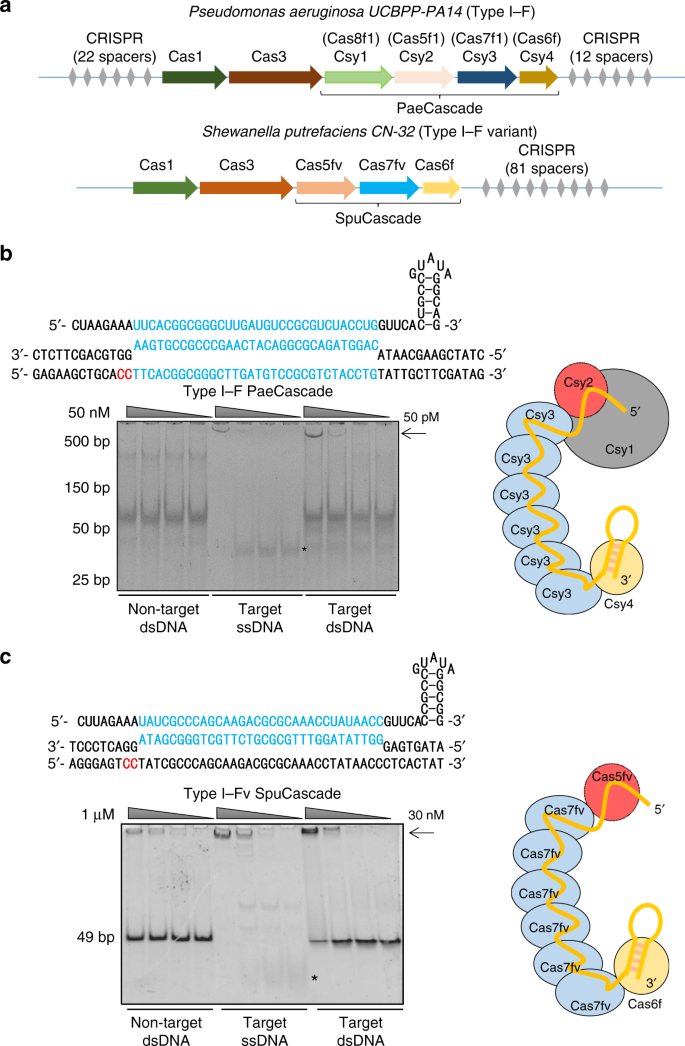


Repurposing Type I F Crispr Cas System As A Transcriptional Activation Tool In Human Cells Nature Communications
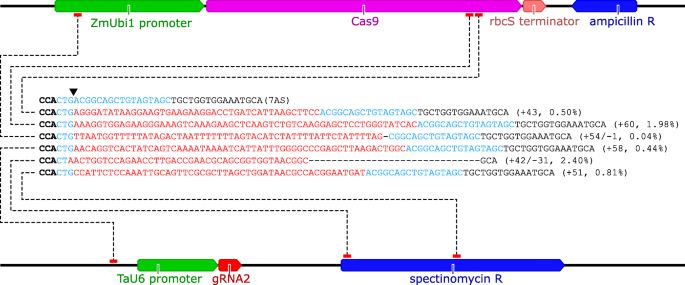


Grna Validation For Wheat Genome Editing With The Crispr Cas9 System Bmc Biotechnology Full Text



Crispr Cas9 Development And Application In Rice Breeding Sciencedirect



Overview Of The Crispr Cas9 Mechanism Of Action A Crispr Cas Download Scientific Diagram



Addgene Crispr Guide
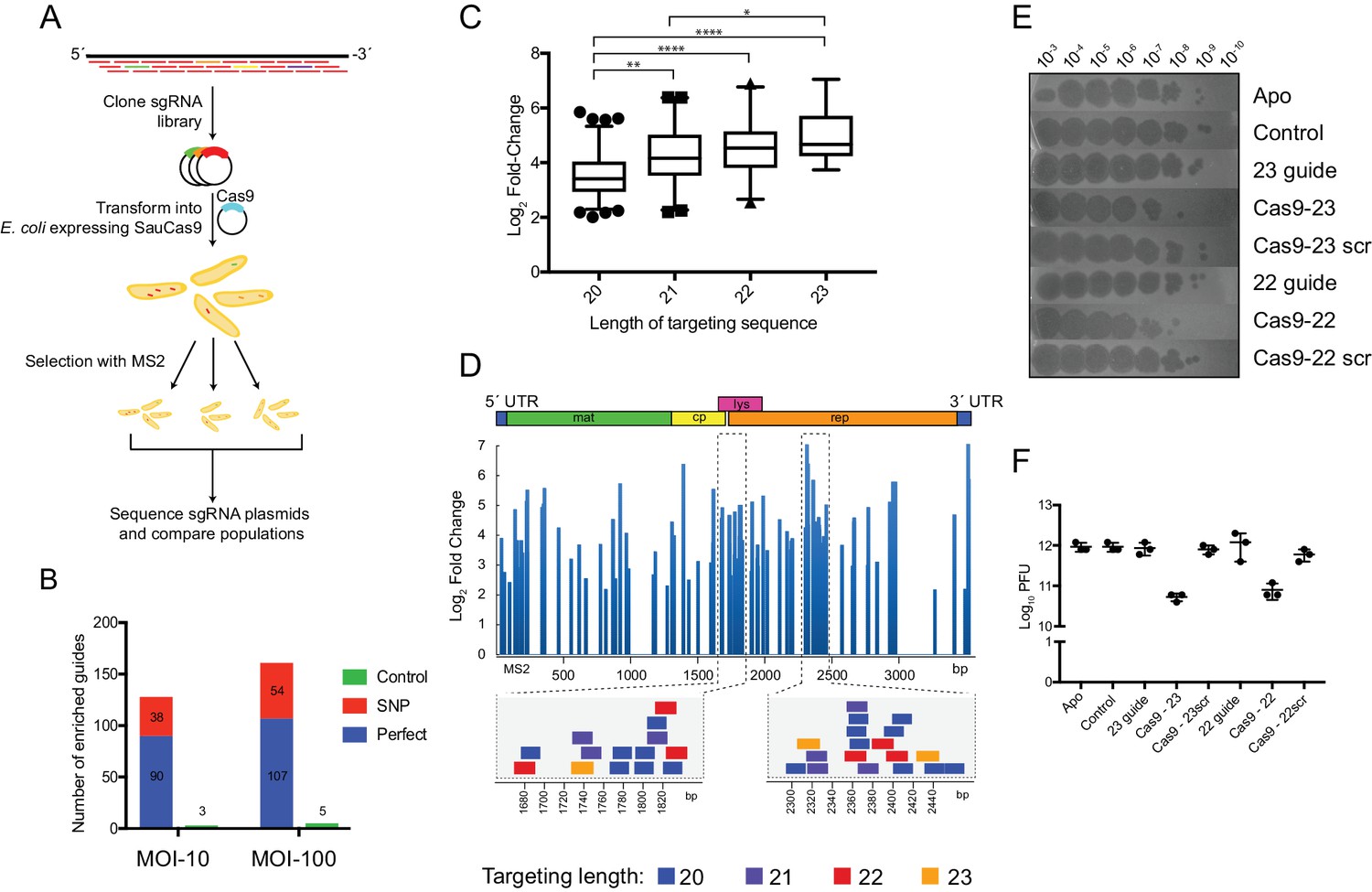


Rna Dependent Rna Targeting By Crispr Cas9 Elife
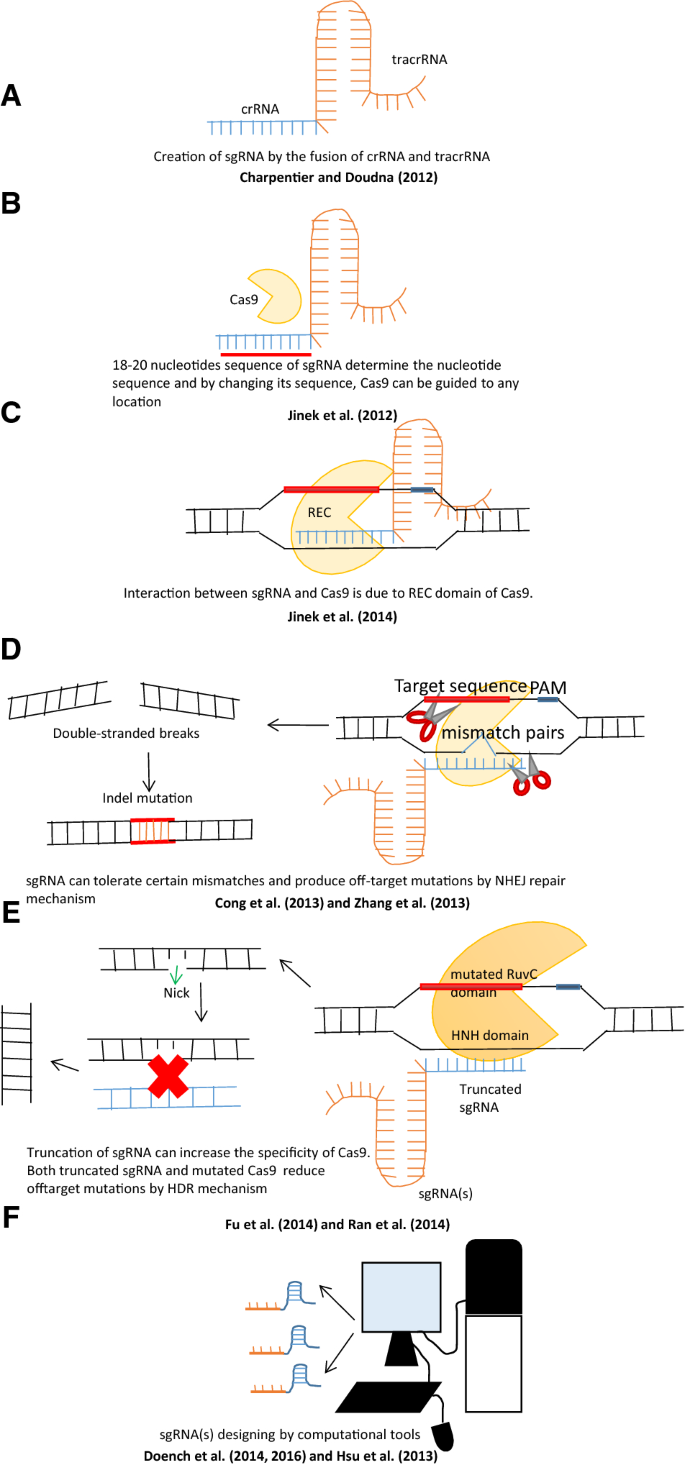


An Overview Of Designing And Selection Of Sgrnas For Precise Genome Editing By The Crispr Cas9 System In Plants Springerlink



Generating Crispr Cas9 Mediated Monoallelic Deletions To Study Enhancer Function In Mouse Embryonic Stem Cells Protocol
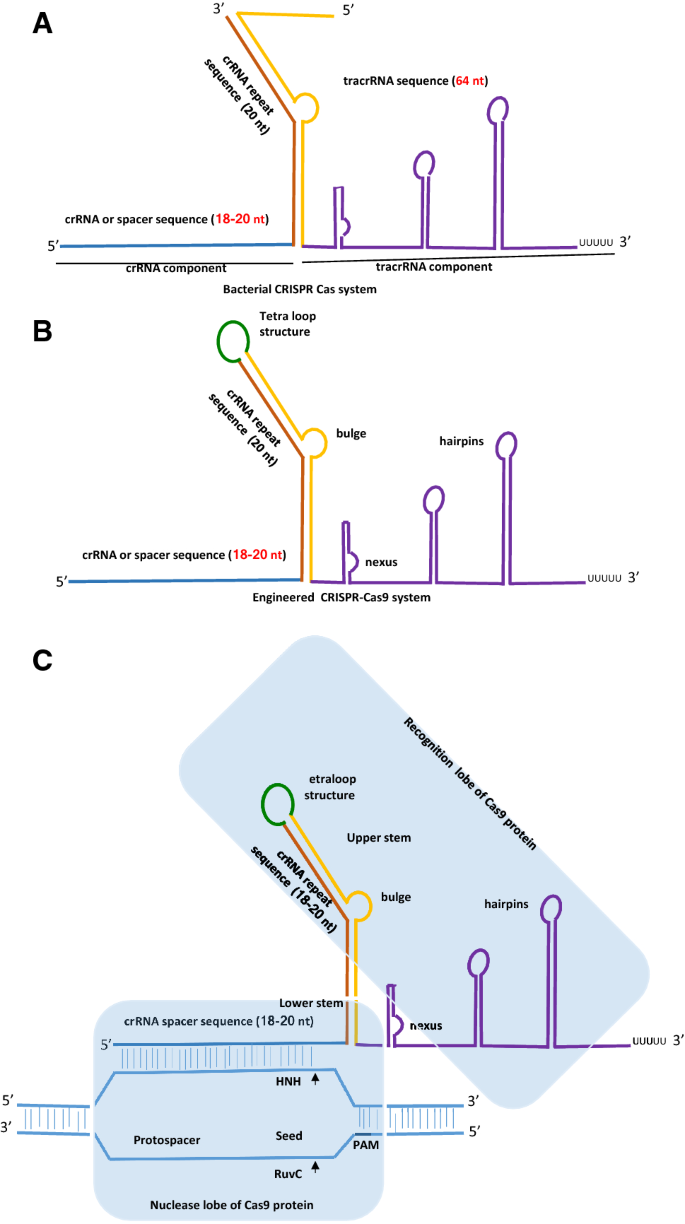


An Overview Of Designing And Selection Of Sgrnas For Precise Genome Editing By The Crispr Cas9 System In Plants Springerlink



No Off Target Mutations In Functional Genome Regions Of A Crispr Cas9 Generated Monkey Model Of Muscular Dystrophy Journal Of Biological Chemistry



Rna Guided Genome Editing In Plants Using A Crispr Cas System Molecular Plant



The Crispr Cas Revolution Reaches The Rna World Cas13 A New Swiss Army Knife For Plant Biologists Wolter 18 The Plant Journal Wiley Online Library
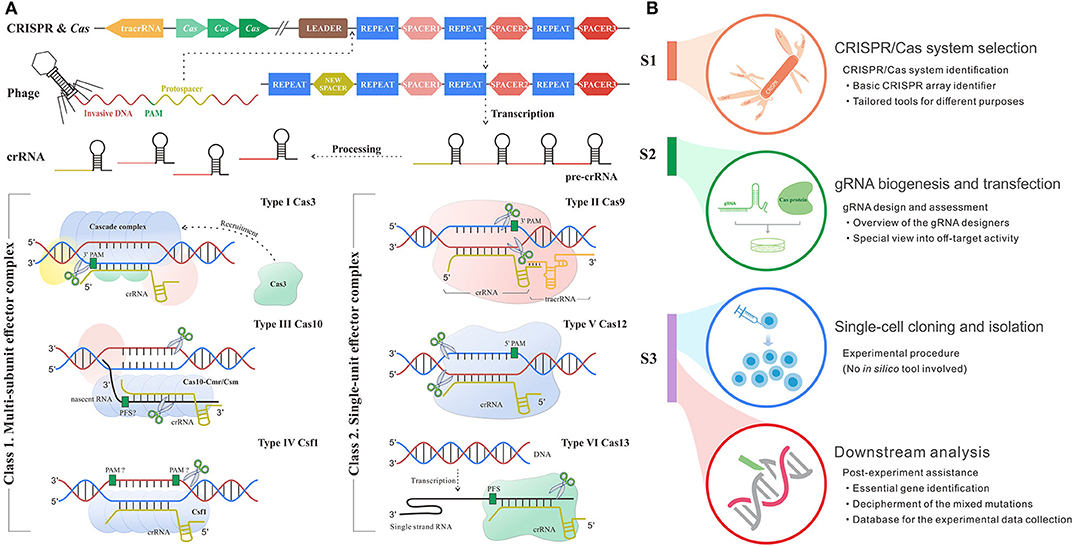


Frontiers In Silico Method In Crispr Cas System An Expedite And Powerful Booster Oncology
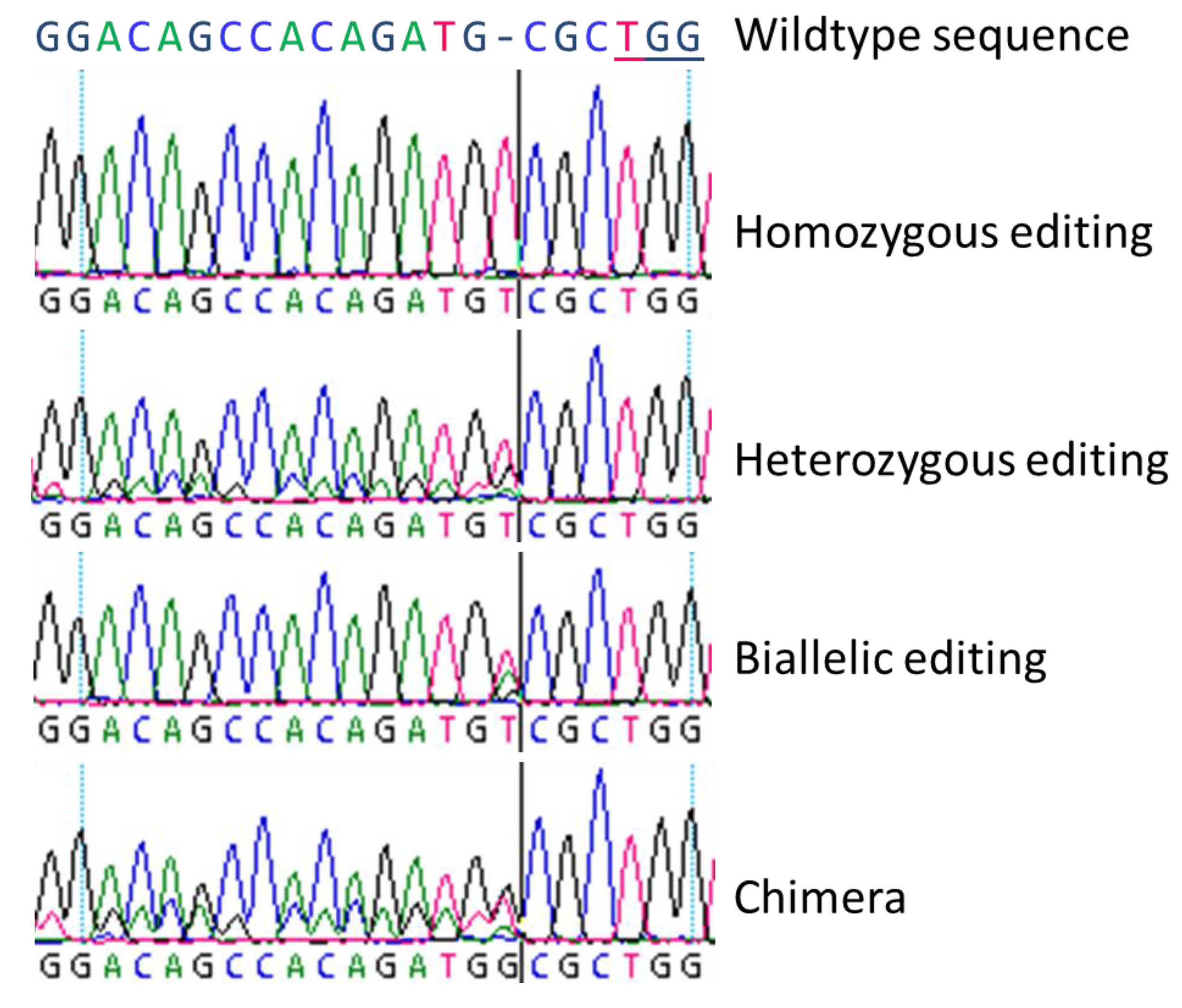


Ijms Free Full Text Evaluating The Efficiency Of Grnas In Crispr Cas9 Mediated Genome Editing In Poplars Html


Crispr Cas9 Targeted Mutagenesis Leads To Simultaneous Modification Of Different Homoeologous Gene Copies In Polyploid Oilseed Rape Brassica Napus Plant Physiology



Addgene Crispr Guide



Figure 3 From Extensive Crispr Rna Modification Reveals Chemical Compatibility And Structure Activity Relationships For Cas9 Biochemical Activity Semantic Scholar
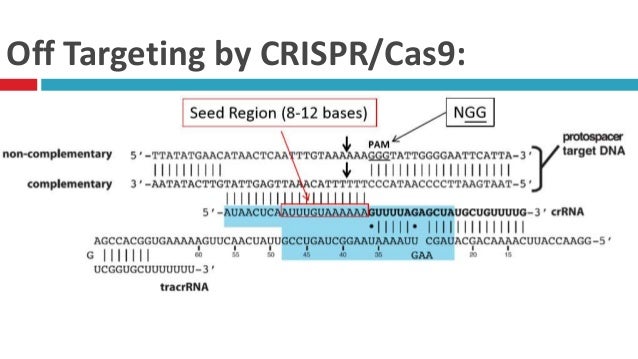


Crispr Cas9


Protospacer Adjacent Motif Pam Distal Sequences Engage Crispr Cas9 Dna Target Cleavage



Off Target Effects In Crispr Cas9 Mediated Genome Engineering Sciencedirect
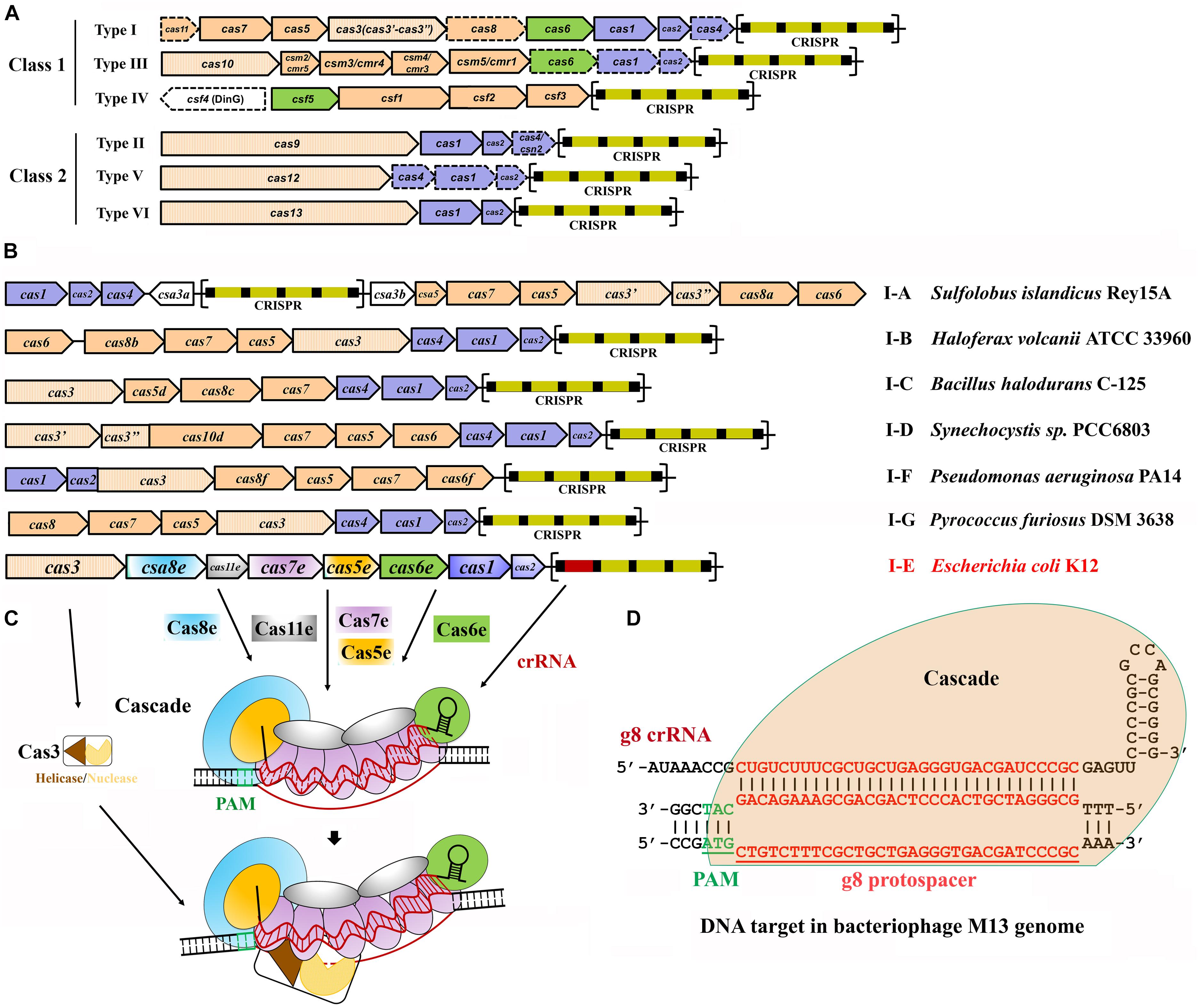


Frontiers Endogenous Type I Crispr Cas From Foreign Dna Defense To Prokaryotic Engineering Bioengineering And Biotechnology



Target Dependent Nickase Activities Of The Crispr Cas Nucleases Cpf1 And Cas9 Abstract Europe Pmc
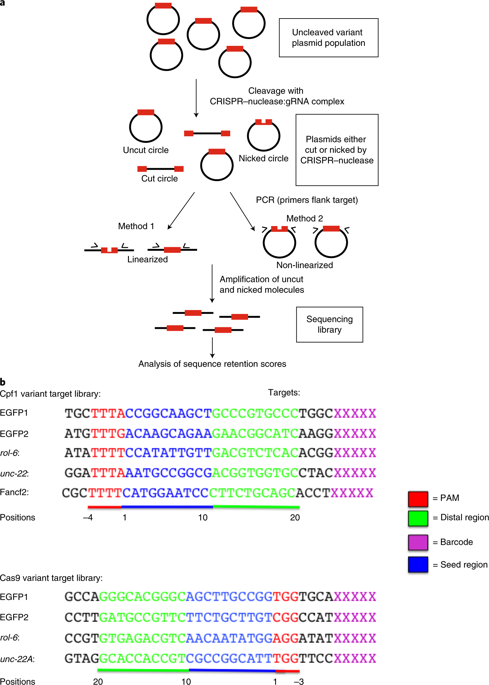


Target Dependent Nickase Activities Of The Crispr Cas Nucleases Cpf1 And Cas9 Nature Microbiology X Mol



In Our Image The Ethics Of Crispr Genome Editing
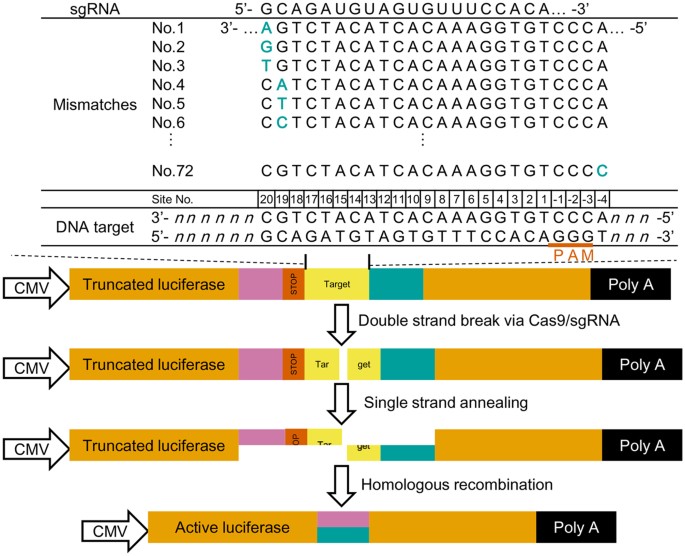


Profiling Single Guide Rna Specificity Reveals A Mismatch Sensitive Core Sequence Scientific Reports



0 件のコメント:
コメントを投稿